This post uses the penguins dataset modified by Allison
Horst in all the code
examples (as an alternative to iris).
library(dplyr)#> Attaching package: 'dplyr'#> The following objects are masked from 'package:stats':#>#> filter, lag#> The following objects are masked from 'package:base':#>#> intersect, setdiff, setequal, unionlibrary(ggplot2)packageVersion("dplyr")#> [1] '1.1.2'
penguins <- palmerpenguins::penguinspenguins
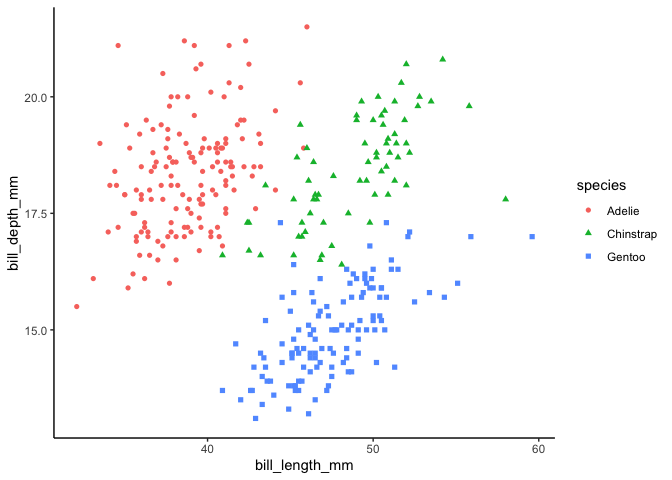
penguins %>%ggplot(aes(bill_length_mm, bill_depth_mm, color = species, shape = species)) +geom_point()#> Warning: Removed 2 rows containing missing values or values outside the scale range#> (`geom_point()`).#> Warning in vp$just: partial match of 'just' to 'justification'
Column-wise Workflows
The new across() function supersedes functionalities of _at, _if,
_all variants. The first argument, .cols, selects the columns you
want to operate on. It uses tidy selection (like select()) so you can
pick variables by position, name, and type. The second argument, .fns,
is a function or list of functions to apply to each column. This can
also be a purrr style formula
penguins_grouped <- penguins %>% group_by(species)penguins_grouped %>%summarize(across(starts_with("bill"), ~ mean(.x, na.rm = TRUE)),n = n())#> species bill_length_mm bill_depth_mm n#> <fct> <dbl> <dbl> <int>#> 1 Adelie 38.8 18.3 152#> 2 Chinstrap 48.8 18.4 68#> 3 Gentoo 47.5 15.0 124
For conditional selection (previous _if variants), predicate function
should be wrapped in where.
# double all numeric columnspenguins %>%mutate(across(where(is.numeric), ~ .x * 2))# count unique values of all character columnspenguins %>%summarize(across(where(is.character), ~ length(unique(.x))))#> # A tibble: 1 × 0
Apply multiple functions using list and use the .names argument to
control column names.
penguins_grouped %>%summarize(across(matches("mm"),list(min = ~ min(.x, na.rm = TRUE),max = ~ max(.x, na.rm = TRUE)),.names = "{fn}_{col}"))#> species min_bill_length_mm max_bill_length_mm min_bill_depth_mm#> <fct> <dbl> <dbl> <dbl>#> 1 Adelie 32.1 46 15.5#> 2 Chinstrap 40.9 58 16.4#> 3 Gentoo 40.9 59.6 13.1#> # ℹ 3 more variables: max_bill_depth_mm <dbl>, min_flipper_length_mm <int>,#> # max_flipper_length_mm <int>
Row-wise Workflows
Row-wise operations require a special type of grouping where each group
consists of a single row. You create this with rowwise().
df <- tibble(student_id = 1:4,test1 = 10:13,test2 = 20:23,test3 = 30:33,test4 = 40:43)df %>% rowwise()#> # Rowwise:#> student_id test1 test2 test3 test4#> <int> <int> <int> <int> <int>#> 1 1 10 20 30 40#> 2 2 11 21 31 41#> 3 3 12 22 32 42#> 4 4 13 23 33 43
rowwise doesn’t need any additional arguments unless you have
variables that identify the rows, like student_id here. This can be
helpful when you want to keep a row identifier.
Like group_by, rowwise doesn’t really do anything itself; it just
changes how the other verbs work.
df %>% mutate(avg = mean(c(test1, test2, test3, test4)))#> student_id test1 test2 test3 test4 avg#> <int> <int> <int> <int> <int> <dbl>#> 1 1 10 20 30 40 26.5#> 2 2 11 21 31 41 26.5#> 3 3 12 22 32 42 26.5#> 4 4 13 23 33 43 26.5df %>%rowwise() %>%mutate(avg = mean(c(test1, test2, test3, test4)))#> # Rowwise:#> student_id test1 test2 test3 test4 avg#> <int> <int> <int> <int> <int> <dbl>#> 1 1 10 20 30 40 25#> 2 2 11 21 31 41 26#> 3 3 12 22 32 42 27#> 4 4 13 23 33 43 28
rowwise takes each row, feeds it into a function, and return a tibble
with the same number of rows. This essentially parallelize a function
over the rows in the dataframe. In this case, the mean() function is
vectorized. But, if a function is already vectorized, then rowwise is
not needed.
df %>% mutate(s = test1 + test2 + test3)#> student_id test1 test2 test3 test4 s#> <int> <int> <int> <int> <int> <int>#> 1 1 10 20 30 40 60#> 2 2 11 21 31 41 63#> 3 3 12 22 32 42 66#> 4 4 13 23 33 43 69df %>%rowwise() %>%mutate(s = test1 + test2 + test3)#> # Rowwise:#> student_id test1 test2 test3 test4 s#> <int> <int> <int> <int> <int> <int>#> 1 1 10 20 30 40 60#> 2 2 11 21 31 41 63#> 3 3 12 22 32 42 66#> 4 4 13 23 33 43 69
Another family of summary functions have “parallel” extensions where you can provide multiple variables in the arguments:
df %>%mutate(min = pmin(test1, test2, test3, test4),max = pmax(test1, test2, test3, test4),string = paste(test1, test2, test3, test4, sep = "-"))#> student_id test1 test2 test3 test4 min max string#> <int> <int> <int> <int> <int> <int> <int> <chr>#> 1 1 10 20 30 40 10 40 10-20-30-40#> 2 2 11 21 31 41 11 41 11-21-31-41#> 3 3 12 22 32 42 12 42 12-22-32-42#> 4 4 13 23 33 43 13 43 13-23-33-43
Where these functions exist, they’ll usually be faster than rowwise.
The advantage of rowwise is that it works with any function, not just
those that are already vectorized.
However, an advantage of rowwise even there is other ways is that it’s
paired with c_across(), which works like c() but uses the same
tidyselect syntax as across(). That makes it easy to operate on
multiple variables:
df %>%rowwise() %>%mutate(min = min(c_across(starts_with("test"))),max = max(c_across(starts_with("test"))))#> # Rowwise:#> student_id test1 test2 test3 test4 min max#> <int> <int> <int> <int> <int> <int> <int>#> 1 1 10 20 30 40 10 40#> 2 2 11 21 31 41 11 41#> 3 3 12 22 32 42 12 42#> 4 4 13 23 33 43 13 43
Plus, a rowwise df will naturally contain exactly the same rows after
summarize(), the same as mutate
df %>%rowwise() %>%summarize(across(starts_with("test"), ~ .x, .names = "{col}_same"))#> test1_same test2_same test3_same test4_same#> <int> <int> <int> <int>#> 1 10 20 30 40#> 2 11 21 31 41#> 3 12 22 32 42#> 4 13 23 33 43
List Columns
Because lists can contain anything, you can use list-columns to keep
related objects together, regardless of what type of thing they are.
List-columns give you a convenient storage mechanism and rowwise gives
you a convenient computation mechanism.
df <- tibble(x = list(1, 2:3, 4:6),y = list(TRUE, 1, "a"),z = list(sum, mean, sd))df#> x y z#> <list> <list> <list>#> 1 <dbl [1]> <lgl [1]> <fn>#> 2 <int [2]> <dbl [1]> <fn>#> 3 <int [3]> <chr [1]> <fn>
df %>%rowwise() %>%summarize(x_length = length(x),y_type = typeof(y),z_call = z(1:5))#> x_length y_type z_call#> <int> <chr> <dbl>#> 1 1 logical 15#> 2 2 double 3#> 3 3 character 1.58
Simulation
The basic idea of using rowwise to perform simulation is to store all
your simulation parameters in a data frame, similar to purrr::pmap.
df <- tribble(~id, ~n, ~min, ~max,1, 3, 0, 1,2, 2, 10, 100,3, 2, 100, 1000,)
Then you can either generate a list-column containing the simulated
values with mutate:
df %>%rowwise() %>%mutate(sim = list(runif(n, min, max)))#> # Rowwise:#> id n min max sim#> <dbl> <dbl> <dbl> <dbl> <list>#> 1 1 3 0 1 <dbl [3]>#> 2 2 2 10 100 <dbl [2]>#> 3 3 2 100 1000 <dbl [2]>
Or taking advantage of summarize’s new features to return multiple
rows per group
df %>%rowwise(everything()) %>%summarize(sim = runif(n, min, max))
In dplyr 1.1, you should use reframe() instead of summarize() to
return multiple rows.
df |>rowwise(everything()) |>reframe(sim = runif(n, min, max))
Without rowwise, you would need to use purrr::pmap to perform the
simulation.
df %>%mutate(sim = purrr::pmap(., ~ runif(..2, ..3, ..4)))#> id n min max sim#> <dbl> <dbl> <dbl> <dbl> <list>#> 1 1 3 0 1 <dbl [3]>#> 2 2 2 10 100 <dbl [2]>#> 3 3 2 100 1000 <dbl [2]>
Group-wise Models
The new nest_by() function works similarly to group_nest()
by_species <- penguins %>% nest_by(species)by_species#> # Rowwise: species#> species data#> <fct> <list<tibble[,7]>>#> 1 Adelie [152 × 7]#> 2 Chinstrap [68 × 7]#> 3 Gentoo [124 × 7]
Now we can use mutate to fit a model to each data frame:
by_species <- by_species %>%rowwise(species) %>%mutate(model = list(lm(bill_length_mm ~ bill_depth_mm, data = data)))by_species#> # Rowwise: species#> species data model#> <fct> <list<tibble[,7]>> <list>#> 1 Adelie [152 × 7] <lm>#> 2 Chinstrap [68 × 7] <lm>#> 3 Gentoo [124 × 7] <lm>
And then extract model summaries or coefficients with summarize() and
broom functions (note that by_species is still a rowwise data
frame):
by_species %>%summarize(broom::glance(model))by_species %>%summarize(broom::tidy(model))
An alternative approach
penguins %>%group_by(species) %>%group_modify(~ broom::tidy(lm(bill_length_mm ~ bill_depth_mm, data = .x)))#> # Groups: species [3]#> species term estimate std.error statistic p.value#> <fct> <chr> <dbl> <dbl> <dbl> <dbl>#> 1 Adelie (Intercept) 23.1 3.03 7.60 3.01e-12#> 2 Adelie bill_depth_mm 0.857 0.165 5.19 6.67e- 7#> 3 Chinstrap (Intercept) 13.4 5.06 2.66 9.92e- 3#> 4 Chinstrap bill_depth_mm 1.92 0.274 7.01 1.53e- 9#> 5 Gentoo (Intercept) 17.2 3.28 5.25 6.60e- 7#> 6 Gentoo bill_depth_mm 2.02 0.219 9.24 1.02e-15
New summarize Features
Use reframe() instead of summarize() to return multiple rows
starting from dplyr 1.1.
Multiple Rows and Columns
Two big changes make summarize() much more flexible. A single summary
expression can now return:
-
A vector of any length, creating multiple rows. (so we can use summary that returns multiple values without
list) -
A data frame, creating multiple columns.
penguins_grouped %>%summarize(bill_length_dist = quantile(bill_length_mm,c(0.25, 0.5, 0.75),na.rm = TRUE),q = c(0.25, 0.5, 0.75))
Or return multiple columns from a single summary expression:
penguins_grouped %>%summarize(tibble(min = min(bill_depth_mm, na.rm = TRUE),max = max(bill_depth_mm, na.rm = TRUE)))#> species min max#> <fct> <dbl> <dbl>#> 1 Adelie 15.5 21.5#> 2 Chinstrap 16.4 20.8#> 3 Gentoo 13.1 17.3
At the first glance this may seem not so different with supplying
multiple name-value pairs. But this can be useful inside functions. For
example, in the previous quantile code it would be nice to be able to
reduce the duplication so that we don’t have to type the quantile values
twice. We can now write a simple function because summary expressions
can now be data frames or tibbles:
quibble <- function(x, q = c(0.25, 0.5, 0.75), na.rm = TRUE) {tibble(x = quantile(x, q, na.rm = na.rm), q = q)}penguins_grouped %>%summarize(quibble(bill_depth_mm))
When combining glue syntax and tidy evaluation, it is easy to dynamically name the column names.
quibble <- function(x, q = c(0.25, 0.5, 0.75), na.rm = TRUE) {tibble("{{ x }}_quantile" := quantile(x, q, na.rm = na.rm),"{{ x }}_q" := q)}penguins_grouped %>%summarize(quibble(flipper_length_mm))
As an aside, if we name the tibble expression in summarize() that part
will be packed in the result, which can be solved by tidyr::unpack.
That’s because when we leave the name off, the data frame result is
automatically unpacked.
penguins_grouped %>%summarize(df = quibble(flipper_length_mm))
Non-summary Context
In combination with rowwise operations, summarize() is now
sufficiently powerful to replace many workflows that previously required
a map() function.
For example, to read all the all the .csv files in the current directory, you could write:
tibble(path = dir(pattern = "\\.csv$")) %>%rowwise(path) %>%summarize(read_csv(path))
Move Columns
New verb relocate is provided to change column positions with the same
syntax as select. The default behavior is to move selected columns to
the left-hand side
penguins %>% relocate(island)penguins %>% relocate(starts_with("bill"))penguins %>% relocate(sex, body_mass_g, .after = species)
Similarly, mutate gains new arguments .after and .before to
control where new columns should appear.
penguins %>%mutate(mass_double = body_mass_g * 2, .before = 1)
Row Mutations
dplyr has a new experimental family of row mutation functions inspired
by SQL’s UPDATE, INSERT, UPSERT, and DELETE. Like the join
functions, they all work with a pair of data frames:
-
rows_update(x, y)updates existing rows in x with values in y. -
rows_patch(x, y)works like rows_update() but only changesNAvalues. -
rows_insert(x, y)adds new rows to x from y. -
rows_upsert(x, y)updates existing rows in x and adds new rows from y. -
rows_delete(x, y)deletes rows in x that match rows in y.
The rows_ functions match x and y using keys. All of them check
that the keys of x and y are valid (i.e. unique) before doing anything.
df <- tibble(a = 1:3, b = letters[c(1:2, NA)], c = 0.5 + 0:2)df#> a b c#> <int> <chr> <dbl>#> 1 1 a 0.5#> 2 2 b 1.5#> 3 3 <NA> 2.5
We can use rows_insert() to add new rows:
new <- tibble(a = c(4, 5), b = c("d", "e"), c = c(3.5, 4.5))rows_insert(df, new)#> # A tibble: 5 × 3#> a b c#> <int> <chr> <dbl>#> 1 1 a 0.5#> 2 2 b 1.5#> 3 3 <NA> 2.5#> 4 4 d 3.5#> 5 5 e 4.5
Note that rows_insert() will fail if we attempt to insert a row that
already exists:
df %>% rows_insert(tibble(a = 3, b = "c"))#> Matching, by = "a"#> Error in `rows_insert()`:#> ! `y` can't contain keys that already exist in `x`.#> ℹ The following rows in `y` have keys that already exist in `x`: `c(1)`.#> ℹ Use `conflict = "ignore"` if you want to ignore these `y` rows.df %>% rows_insert(tibble(a = 3, b = "c"), by = c("a", "b"))#> a b c#> <int> <chr> <dbl>#> 1 1 a 0.5#> 2 2 b 1.5#> 3 3 <NA> 2.5#> 4 3 c NA
If you want to update existing values, use rows_update(). It will
throw an error if one of the rows to update does not exist:
df %>% rows_update(tibble(a = 3, b = "c"))#> # A tibble: 3 × 3#> a b c#> <int> <chr> <dbl>#> 1 1 a 0.5#> 2 2 b 1.5#> 3 3 c 2.5df %>% rows_update(tibble(a = 4, b = "d"))#> Matching, by = "a"#> Error in `rows_update()`:#> ! `y` must contain keys that already exist in `x`.#> ℹ The following rows in `y` have keys that don't exist in `x`: `c(1)`.#> ℹ Use `unmatched = "ignore"` if you want to ignore these `y` rows.
rows_patch() is a variant of rows_update() that will only update
values in x that are NA.
df %>% rows_patch(tibble(a = 1:3, b = "patch"))#> # A tibble: 3 × 3#> a b c#> <int> <chr> <dbl>#> 1 1 a 0.5#> 2 2 b 1.5#> 3 3 patch 2.5
row_upsert update a df or insert new rows.
df %>%rows_upsert(tibble(a = 3, b = "c")) %>% # updaterows_upsert(tibble(a = 4, b = "d")) # insert#> Matching, by = "a"#> # A tibble: 4 × 3#> a b c#> <int> <chr> <dbl>#> 1 1 a 0.5#> 2 2 b 1.5#> 3 3 c 2.5#> 4 4 d NA
Context Dependent Expressions
n() is a special function in dplyr which return the number of
observations in the current group. Now the new version comes with more
such special functions, aka context dependent expressions. These
functions return information about the “current” group or “current”
variable, so only work inside specific contexts like summarize() and
mutate(). Specifically, a family of cur_ functions are added:
-
cur_data()gives the current data for the current group (excluding grouping variables,cur_data_allin developmental version returns grouping variables as well) -
cur_group()gives the group keys, a tibble with one row and one column for each grouping variable. -
cur_group_id()gives a unique numeric identifier for the current group -
cur_column()gives the name of the current column (inacross()only).
cur_data() is deprecated in favor of pick(col1, col2, ...) in dplyr
1.1.
df <- tibble(g = sample(rep(letters[1:3], 1:3)),x = runif(6),y = runif(6))gf <- df %>% group_by(g)gf %>% reframe(row = cur_group_rows())#> g row#> <chr> <int>#> 1 a 4#> 2 b 5#> 3 b 6#> 4 c 1#> 5 c 2#> 6 c 3gf %>% reframe(data = list(cur_group()))#> # A tibble: 3 × 2#> g data#> <chr> <list>#> 1 a <tibble [1 × 1]>#> 2 b <tibble [1 × 1]>#> 3 c <tibble [1 × 1]>gf %>% reframe(data = list(pick(everything())))#> g data#> <chr> <list>#> 1 a <tibble [1 × 2]>#> 2 b <tibble [2 × 2]>#> 3 c <tibble [3 × 2]># cur_column() is not related to groupsgf %>% mutate(across(everything(), ~ paste(cur_column(), round(.x, 2))))#> # Groups: g [3]#> g x y#> <chr> <chr> <chr>#> 1 c x 0.84 y 0.59#> 2 c x 0.56 y 0.52#> 3 c x 0.08 y 0.16#> 4 a x 0.96 y 0.28#> 5 b x 0.02 y 0.6#> 6 b x 0.32 y 0.1
Superseded Functions
top_n(), sample_n(), and sample_frac() have been superseded in
favor of a new family of slice helpers: slice_min(), slice_max(),
slice_head(), slice_tail(), slice_random().
# select penguins per group on body masspenguins_grouped %>%slice_max(body_mass_g, n = 1)#> # Groups: species [3]#> species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g#> <fct> <fct> <dbl> <dbl> <int> <int>#> 1 Adelie Biscoe 43.2 19 197 4775#> 2 Chinstrap Dream 52 20.7 210 4800#> 3 Gentoo Biscoe 49.2 15.2 221 6300#> # ℹ 2 more variables: sex <fct>, year <int>penguins_grouped %>%slice_min(body_mass_g, n = 1)#> # Groups: species [3]#> species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g#> <fct> <fct> <dbl> <dbl> <int> <int>#> 1 Adelie Biscoe 36.5 16.6 181 2850#> 2 Adelie Biscoe 36.4 17.1 184 2850#> 3 Chinstrap Dream 46.9 16.6 192 2700#> 4 Gentoo Biscoe 42.7 13.7 208 3950#> # ℹ 2 more variables: sex <fct>, year <int>
# random samplingpenguins %>%slice_sample(n = 10)penguins %>%slice_sample(prop = 0.1)
summarize() gains new argument .groups to control grouping structure
of theh result.
-
.groups = "drop_last"drops the last grouping level (i.e. the default behaviour). -
.groups = "drop"drops all grouping levels and returns a tibble. -
.groups = "keep"preserves the grouping of the input. -
.groups = "rowwise"turns each row into its own group.
Other Changes
The new rename_with() makes it easier to rename variables
programmatically:
penguins %>%rename_with(stringr::str_to_upper)
You can optionally choose which columns to apply the transformation to with the second argument:
penguins %>%rename_with(stringr::str_to_upper, starts_with("bill"))
mutate() gains argument .keep that allows you to control which
columns are retained in the output:
penguins %>% mutate(double_mass = body_mass_g * 2,island_lower = stringr::str_to_lower(island),.keep = "used")penguins %>% mutate(double_mass = body_mass_g * 2, .keep = "none")
Recipes
This in-progress section documents tasks that would otherwise been impossible or laborious with previous version of dplyr.
Replace Missing Values in Multiple Columnes
Since tidyr::replace_na does not support tidy select syntax, replacing
NA values in multiple columns could be a drudgery. Now this is made easy
with coalesce and across
penguins %>% summarize(across(starts_with("bill"), ~ sum(is.na(.x))))#> # A tibble: 1 × 2#> bill_length_mm bill_depth_mm#> <int> <int>#> 1 2 2penguins %>%mutate(across(starts_with("bill"), ~ coalesce(.x, 0))) %>%summarize(across(starts_with("bill"), ~ sum(is.na(.x))))#> # A tibble: 1 × 2#> bill_length_mm bill_depth_mm#> <int> <int>#> 1 0 0
Rolling Regression
We can easily perform rolling computation with the slider package and
pick().
library(slider)library(lubridate)#>#> Attaching package: 'lubridate'#> The following objects are masked from 'package:base':#>#> date, intersect, setdiff, union# historical stock prices from 2014-2018 for Google, Amazon, Facebook and Applestock <- tsibbledata::gafa_stock %>% select(Symbol, Date, Close, Volume)stock
# Arrange and group by `Symbol` (i.e. Google)stock <- stock %>%arrange(Symbol, Date) %>%group_by(Symbol)linear_model <- function(df) {lm(Close ~ Volume, data = df)}# 10 day rolling regression per groupstock %>%mutate(model = slide_index(pick(Close, Volume),Date,linear_model,.before = days(9),.complete = TRUE))